Immune Response of Plants – Part 2
22.09.2017 / Scienceandmore / Category: Plant Biology
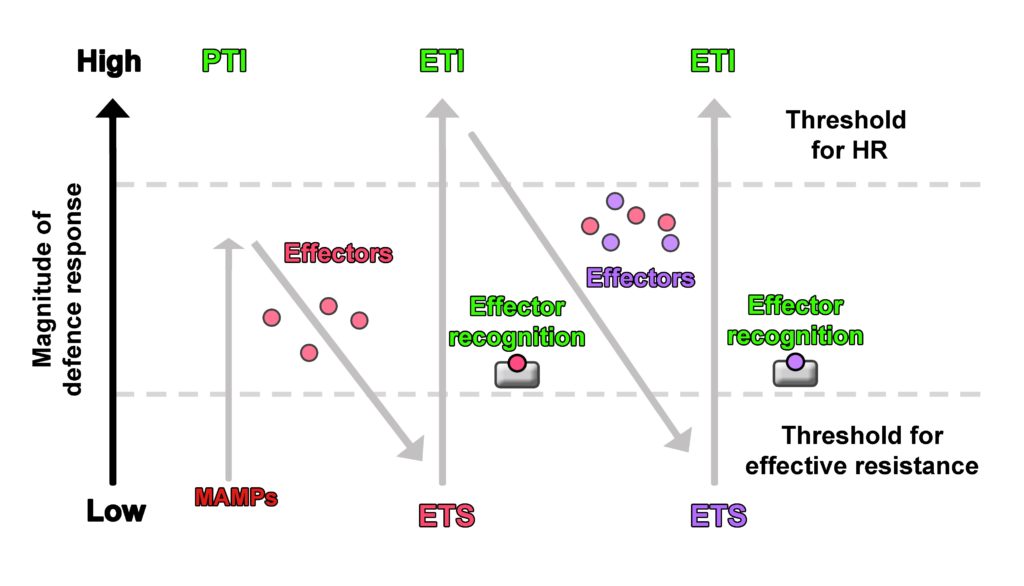
Recently, the zig zag model for plant-pathogen interaction was criticised since it is based on the plant’s interaction with biotrophic microbes, i.e. pathogens that keep their host plans alive while feeding on them, and lacked integration of symbiotic interactions, or the response to necrotrophic pathogens. Hypersensitive response (HR), a form of programmed cell death, has been suggested as a major immune response of the plant in the zig zag model (1), and research suggests that it is effective against biotrophic microbes (2). In terms of an infection with necrotrophic pathogens, HR would, however, rather be a favourable result, since necrotrophic pathogens aim to kill plant cells in order to obtain nutrients. HR would therefore contribute to an invasion by necrotrophs and be counterproductive for the plant.
Also, the environmental context such as the plant’s previous encounters with pathogens, presence of beneficial microbes, etc. have not been considered in the zig zag model. In this model, inducible plant immune response is defined as a mechanism in which perception of MAMPs and microbial effectors result in immunity (PTI/ETI) or susceptibility (ETS). Instead, it was suggested that the plant immune response should rather be defined as a mechanism that integrates various signals and results in different outcomes which cannot simply be categorised into ETI, PTS and PTI (3). An alternative model to describe inducible plant immune response has been proposed by Cook et al. (4) with the invasion model. In this model, compounds that elicit the plant immune response are not categorised into the strict categories of MAMPs or effectors but into invasion patterns (IPs). These IPs are not defined from the point of perception and response by the host plant, but from the point of their main function in the microorganism or plant. IPs are for example microbial-derived compounds such as the fungal chitin and bacterial flagellin, which would have been described as MAMPs in the zig zag model, but also host plant-derived modified-self compounds that indicate an invasion. Modified-self compounds are for example damage associated molecular patterns (DAMPs) such as components of the exterior of the cell that are released after wounding. Well described DAMPs are pectin fragments that are released from the cell wall after penetration by a fungus for instance. However, the main function of pectin is in plant cell wall structure and not as indicator of invasion. DAMPs play an important role in plant immune responses but have not been integrated in the zig zag model (3).
Also, a classification of immune response eliciting compounds into MAMPs or effectors is objected to in the invasion model, since some of these compounds have been found to not fit into the categories of MAMPs and effectors (3).
Contrary to the zig zag model, in the invasion model the host plant’s responses are not classified into PTI or ETI, but into IP triggered responses (IPTRs). The difference is that perception of IPs (by invasion pattern receptors (IPRs)) results in a) continued symbiosis between invader and host plant, or b) end of symbiosis of invader and host plant; instead of immunity (PTI/ETI) or susceptibility (ETS) as conceptualised in the zig zag model. These outcomes of continuation or discontinuation of symbiosis are determined by the overall effect of elicited host plant responses (that can be synergistic and/or antagonistic) (4).
From the invader perspective, mechanisms that lead to a) the failure to supress IPTR, b) the suppression of IPTR (for example by biotrophic invaders), or c) the utilisation of IPTR (for example by necrotrophic invaders to drive cell death) determine if the symbiosis with the host plant is either continued or discontinued. Invaders may deploy IPs to influence the outcome of the symbiosis by manipulating IPTRs. When these IPs are used or the symbiosis is continued, further IPs could be released such as components that indicate a modification of host plant processes or DAMPs that in turn could be perceived by IPRs and lead to a discontinuation or continuation of the symbiosis, depending on the triggered plant responses (4).
In the zig zag model, plant immune response and resistance are associated with HR, which, however, would rather result in a susceptibility to necrotrophic pathogens instead of resistance. In the invasion model, infection by necrotrophs leads to the release of DAMPs such as pectin fragments from the cell wall, resulting in triggering of appropriate defence responses. Additionally, pro-death mechanisms utilised by necrotrophic microorganisms such as specific toxins to hijack the plants immune response to drive HR for instance did not really fit in the zig zag model but are integrated in the invasion model.
Symbiosis of host plants with beneficial microbes also depends on IPs (effectors) that suppress the plants defence response, similar to interactions with pathogens. Arbuscular mycorrhiza fungi, which are fungi that colonise the roots of a wide range of land plants and that are essential for the host plants development, growth and stress tolerance (5), also deploy effector to attenuate the plants immune response. This, however, does not represent susceptibility of the host plant (ETS in the zig zag model) in a narrow sense and, therefore, does not really fit into the zig zag model. In the invasion model on the other hand, arbuscular mycorrhiza fungi deploy IPs that suppress IPTRs, which otherwise would lead to a discontinuation of symbiosis (4).
Finally, it has to be pointed out that both the zig zag model and the invasion model are models after all that try to incorporate as many aspects of plant-microbe interaction and make sense of them. Nevertheless, models are generalisations and are therefore incomplete but form the basis for new scientific hypotheses and progress.
References
1. Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323-9.
2. Coll, N.S., Epple, P., Dangl, J.L. (2011): Programmed cell death in the plant immune system. Cell Death and Differentiation, 18, 1247-56.
3. Pritchard, L. and Birch, P. R. J. (2014) The zigzag model of plant–microbe interactions: is it time to move on? Mol Plant Pathol. 15(9), 865-70.
4. Cook, D.E., Mesarich, C.H., Thomma, B.P. (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol. 53, 541-63.
5. Harrier, L.A (2001) The arbuscular mycorrhizal symbiosis: a molecular review of the fungal dimension. Journal of Experimental Botany, 52(1): 469-78.